Sensitivity Analysis for Measurement Error
2024-07-01
Source:vignettes/measurement_error.Rmd
measurement_error.RmdThis example demonstrates how to conduct sensitivity analysis for
measurement error by using cmsens. For this purpose, we
simulate some data containing a continuous baseline confounder \(C_1\), a binary baseline confounder \(C_2\), a binary exposure \(A\), a binary mediator \(M\) and a binary outcome \(Y\). The true regression models for \(A\), \(M\)
and \(Y\) are: \[logit(E(A|C_1,C_2))=0.2+0.5C_1+0.1C_2\]
\[logit(E(M|A,C_1,C_2))=1+2A+1.5C_1+0.8C_2\]
\[logit(E(Y|A,M,C_1,C_2)))=-3-0.4A-1.2M+0.5AM+0.3C_1-0.6C_2\]
Then, we generate some measurement errors for \(C_1\) and \(A\).
set.seed(1)
expit <- function(x) exp(x)/(1+exp(x))
n <- 10000
C1 <- rnorm(n, mean = 1, sd = 0.5)
C1_error <- C1 + rnorm(n, 0, 0.05)
C2 <- rbinom(n, 1, 0.6)
A <- rbinom(n, 1, expit(0.2 + 0.5*C1 + 0.1*C2))
mc <- matrix(c(0.9,0.1,0.1,0.9), nrow = 2)
A_error <- A
for (j in 1:2) {
A_error[which(A_error == c(0,1)[j])] <-
sample(x = c(0,1), size = length(which(A_error == c(0,1)[j])),
prob = mc[, j], replace = TRUE)
}
M <- rbinom(n, 1, expit(1 + 2*A + 1.5*C1 + 0.8*C2))
Y <- rbinom(n, 1, expit(-3 - 0.4*A - 1.2*M + 0.5*A*M + 0.3*C1 - 0.6*C2))
data <- data.frame(A, A_error, M, Y, C1, C1_error, C2)The DAG for this scientific setting is:
## Registered S3 methods overwritten by 'lme4':
## method from
## cooks.distance.influence.merMod car
## influence.merMod car
## dfbeta.influence.merMod car
## dfbetas.influence.merMod car
cmdag(outcome = "Y", exposure = "A", mediator = "M",
basec = c("C1", "C2"), postc = NULL, node = TRUE, text_col = "white")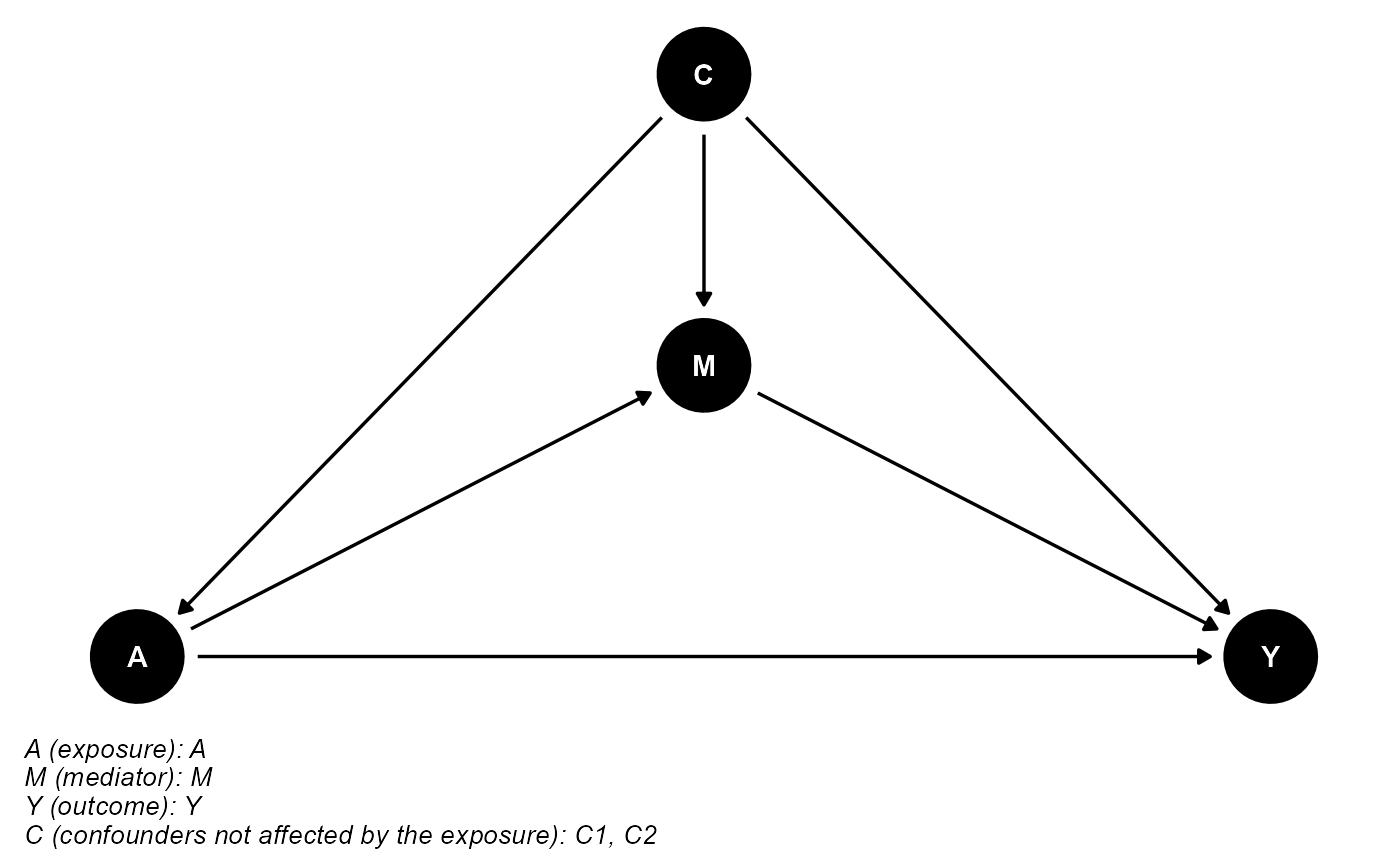
A Continuous Variable Measured with Error
Firstly, we assume \(C1\) was measured with error. \(C_1\) is continuous, so the measurement error can be corrected by regression calibration or SIMEX. We use the regression-based approach for illustration. The naive results obtained by fitting data with measurement error:
res_naive_cont <- cmest(data = data, model = "rb", outcome = "Y", exposure = "A",
mediator = "M", basec = c("C1_error", "C2"), EMint = TRUE,
mreg = list("logistic"), yreg = "logistic",
astar = 0, a = 1, mval = list(1),
estimation = "paramfunc", inference = "delta")
summary(res_naive_cont)## Causal Mediation Analysis
##
## # Outcome regression:
##
## Call:
## glm(formula = Y ~ A + M + A * M + C1_error + C2, family = binomial(),
## data = getCall(x$reg.output$yreg)$data, weights = getCall(x$reg.output$yreg)$weights)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -0.5658 -0.2097 -0.1763 -0.1527 3.1992
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -2.5688 0.2608 -9.852 < 2e-16 ***
## A -0.8882 0.7581 -1.172 0.24139
## M -1.8387 0.2858 -6.434 1.24e-10 ***
## C1_error 0.4856 0.1481 3.280 0.00104 **
## C2 -0.6142 0.1500 -4.094 4.24e-05 ***
## A:M 1.1063 0.7790 1.420 0.15555
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 1858.7 on 9999 degrees of freedom
## Residual deviance: 1799.6 on 9994 degrees of freedom
## AIC: 1811.6
##
## Number of Fisher Scoring iterations: 7
##
##
## # Mediator regressions:
##
## Call:
## glm(formula = M ~ A + C1_error + C2, family = binomial(), data = getCall(x$reg.output$mreg[[1L]])$data,
## weights = getCall(x$reg.output$mreg[[1L]])$weights)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -3.4202 0.0948 0.1395 0.2539 1.0725
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 0.9916 0.1160 8.552 < 2e-16 ***
## A 2.1481 0.1473 14.586 < 2e-16 ***
## C1_error 1.4448 0.1193 12.109 < 2e-16 ***
## C2 0.6769 0.1193 5.673 1.4e-08 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 2825.7 on 9999 degrees of freedom
## Residual deviance: 2306.7 on 9996 degrees of freedom
## AIC: 2314.7
##
## Number of Fisher Scoring iterations: 7
##
##
## # Effect decomposition on the odds ratio scale via the regression-based approach
##
## Closed-form parameter function estimation with
## delta method standard errors, confidence intervals and p-values
##
## Estimate Std.error 95% CIL 95% CIU P.val
## Rcde 1.24383 0.22304 0.87524 1.768 0.223674
## Rpnde 1.01905 0.17645 0.72578 1.431 0.913235
## Rtnde 1.20938 0.21034 0.86004 1.701 0.274370
## Rpnie 0.80077 0.05414 0.70138 0.914 0.001017 **
## Rtnie 0.95034 0.06599 0.82942 1.089 0.463197
## Rte 0.96844 0.15560 0.70683 1.327 0.841789
## ERcde 0.18846 0.16640 -0.13769 0.515 0.257410
## ERintref -0.16941 0.09996 -0.36533 0.027 0.090128 .
## ERintmed 0.14862 0.08786 -0.02358 0.321 0.090726 .
## ERpnie -0.19923 0.05414 -0.30535 -0.093 0.000234 ***
## ERcde(prop) -5.97114 34.38901 -73.37236 61.430 0.862152
## ERintref(prop) 5.36768 26.42572 -46.42577 57.161 0.839039
## ERintmed(prop) -4.70891 23.17884 -50.13861 40.721 0.839013
## ERpnie(prop) 6.31237 31.15632 -54.75290 67.378 0.839445
## pm 1.60346 8.40147 -14.86311 18.070 0.848639
## int 0.65876 3.24847 -5.70811 7.026 0.839298
## pe 6.97114 34.38901 -60.43008 74.372 0.839359
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Rcde: controlled direct effect odds ratio; Rpnde: pure natural direct effect odds ratio; Rtnde: total natural direct effect odds ratio; Rpnie: pure natural indirect effect odds ratio; Rtnie: total natural indirect effect odds ratio; Rte: total effect odds ratio; ERcde: excess relative risk due to controlled direct effect; ERintref: excess relative risk due to reference interaction; ERintmed: excess relative risk due to mediated interaction; ERpnie: excess relative risk due to pure natural indirect effect; ERcde(prop): proportion ERcde; ERintref(prop): proportion ERintref; ERintmed(prop): proportion ERintmed; ERpnie(prop): proportion ERpnie; pm: overall proportion mediated; int: overall proportion attributable to interaction; pe: overall proportion eliminated)
##
## Relevant variable values:
## $a
## [1] 1
##
## $astar
## [1] 0
##
## $yval
## [1] "1"
##
## $mval
## $mval[[1]]
## [1] 1
##
##
## $basecval
## $basecval[[1]]
## [1] 0.996522
##
## $basecval[[2]]
## [1] 0.5936The results corrected by regression calibration:
res_rc_cont <- cmsens(object = res_naive_cont, sens = "me", MEmethod = "rc",
MEvariable = "C1_error", MEvartype = "con", MEerror = 0.05)
summary(res_rc_cont)## Sensitivity Analysis For Measurement Error
##
## The variable measured with error: C1_error
## Type of the variable measured with error: continuous
##
## # Measurement error 1:
## [1] 0.05
##
## ## Error-corrected regressions for measurement error 1:
##
## ### Outcome regression:
## Call:
## rcreg(reg = getCall(x$sens[[1L]]$reg.output$yreg)$reg, formula = Y ~
## A + M + A * M + C1_error + C2, data = getCall(x$sens[[1L]]$reg.output$yreg)$data,
## MEvariable = "C1_error", MEerror = 0.05, variance = TRUE,
## nboot = 400, weights = getCall(x$sens[[1L]]$reg.output$yreg)$weights)
##
## Naive coefficient estimates:
## (Intercept) A M C1_error C2 A:M
## -2.5688363 -0.8881518 -1.8387439 0.4856104 -0.6142071 1.1063464
##
## Naive var-cov estimates:
## (Intercept) A M C1_error C2
## (Intercept) 0.067990581 -0.0545603114 -0.048204212 -0.0154891419 -0.0071194416
## A -0.054560311 0.5747507824 0.054868704 -0.0004875592 0.0004879114
## M -0.048204212 0.0548687045 0.081668670 -0.0080216586 -0.0027107101
## C1_error -0.015489142 -0.0004875592 -0.008021659 0.0219233192 -0.0001013951
## C2 -0.007119442 0.0004879114 -0.002710710 -0.0001013951 0.0225065272
## A:M 0.055848645 -0.5747208888 -0.077864595 -0.0011835634 -0.0008171310
## A:M
## (Intercept) 0.055848645
## A -0.574720889
## M -0.077864595
## C1_error -0.001183563
## C2 -0.000817131
## A:M 0.606845225
##
## Variable measured with error:
## C1_error
## Measurement error:
## 0.05
## Error-corrected results:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -2.5717 0.2542 -10.116 < 2e-16 ***
## A -0.8886 4.4880 -0.198 0.84306
## M -1.8405 0.2816 -6.535 6.65e-11 ***
## C1_error 0.4905 0.1456 3.368 0.00076 ***
## C2 -0.6142 0.1495 -4.108 4.03e-05 ***
## A:M 1.1063 4.5006 0.246 0.80583
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## ### Mediator regressions:
## Call:
## rcreg(reg = getCall(x$sens[[1L]]$reg.output$mreg[[1L]])$reg,
## formula = M ~ A + C1_error + C2, data = getCall(x$sens[[1L]]$reg.output$mreg[[1L]])$data,
## MEvariable = "C1_error", MEerror = 0.05, variance = TRUE,
## nboot = 400, weights = getCall(x$sens[[1L]]$reg.output$mreg[[1L]])$weights)
##
## Naive coefficient estimates:
## (Intercept) A C1_error C2
## 0.9915629 2.1481051 1.4448263 0.6769050
##
## Naive var-cov estimates:
## (Intercept) A C1_error C2
## (Intercept) 0.013444822 -0.0040002083 -0.0092978270 -0.0070380421
## A -0.004000208 0.0216896845 -0.0008364503 0.0002395075
## C1_error -0.009297827 -0.0008364503 0.0142375505 0.0008566472
## C2 -0.007038042 0.0002395075 0.0008566472 0.0142371683
##
## Variable measured with error:
## C1_error
## Measurement error:
## 0.05
## Error-corrected results:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.9784 0.1208 8.101 6.10e-16 ***
## A 2.1465 0.1509 14.229 < 2e-16 ***
## C1_error 1.4591 0.1274 11.454 < 2e-16 ***
## C2 0.6769 0.1208 5.602 2.18e-08 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
##
## ## Error-corrected causal effects on the risk ratio scale for measurement error 1:
## Estimate Std.error 95% CIL 95% CIU P.val
## Rcde 1.24332 0.21790 0.88187 1.753 0.214000
## Rpnde 1.01851 0.51560 0.37762 2.747 0.971104
## Rtnde 1.20880 0.21487 0.85320 1.713 0.286051
## Rpnie 0.80064 0.05377 0.70189 0.913 0.000931 ***
## Rtnie 0.95024 0.40376 0.41320 2.185 0.904375
## Rte 0.96782 0.15935 0.70088 1.336 0.842532
## ERcde 0.18801 0.16191 -0.12932 0.505 0.245553
## ERintref -0.16951 0.50414 -1.15760 0.819 0.736699
## ERintmed 0.14867 0.44221 -0.71805 1.015 0.736722
## ERpnie -0.19936 0.05377 -0.30475 -0.094 0.000209 ***
## ERcde(prop) -5.84268 33.30469 -71.11868 59.433 0.860741
## ERintref(prop) 5.26756 25.15247 -44.03038 54.566 0.834116
## ERintmed(prop) -4.62011 22.05610 -47.84926 38.609 0.834081
## ERpnie(prop) 6.19523 30.72226 -54.01929 66.410 0.840188
## pm 1.57512 17.92074 -33.54889 36.699 0.929961
## int 0.64746 3.09806 -5.42463 6.720 0.834458
## pe 6.84268 33.30469 -58.43331 72.119 0.837215
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## ----------------------------------------------------------------
##
## (Rcde: controlled direct effect risk ratio; Rpnde: pure natural direct effect risk ratio; Rtnde: total natural direct effect risk ratio; Rpnie: pure natural indirect effect risk ratio; Rtnie: total natural indirect effect risk ratio; Rte: total effect risk ratio; ERcde: excess relative risk due to controlled direct effect; ERintref: excess relative risk due to reference interaction; ERintmed: excess relative risk due to mediated interaction; ERpnie: excess relative risk due to pure natural indirect effect; ERcde(prop): proportion ERcde; ERintref(prop): proportion ERintref; ERintmed(prop): proportion ERintmed; ERpnie(prop): proportion ERpnie; pm: overall proportion mediated; int: overall proportion attributable to interaction; pe: overall proportion eliminated)
##
## Relevant variable values:
## $a
## [1] 1
##
## $astar
## [1] 0
##
## $yval
## [1] "1"
##
## $mval
## $mval[[1]]
## [1] 1
##
##
## $basecval
## $basecval[[1]]
## [1] 0.996522
##
## $basecval[[2]]
## [1] 0.5936The results corrected by SIMEX:
res_simex_cont <- cmsens(object = res_naive_cont, sens = "me", MEmethod = "simex",
MEvariable = "C1_error", MEvartype = "con", MEerror = 0.05)
summary(res_simex_cont)## Sensitivity Analysis For Measurement Error
##
## The variable measured with error: C1_error
## Type of the variable measured with error: continuous
##
## # Measurement error 1:
## [1] 0.05
##
## ## Error-corrected regressions for measurement error 1:
##
## ### Outcome regression:
## Call:
## simexreg(reg = getCall(x$sens[[1L]]$reg.output$yreg)$reg, formula = Y ~
## A + M + A * M + C1_error + C2, data = getCall(x$sens[[1L]]$reg.output$yreg)$data,
## MEvariable = "C1_error", MEvartype = "continuous", MEerror = 0.05,
## variance = TRUE, lambda = c(0.5, 1, 1.5, 2), B = 200, weights = getCall(x$sens[[1L]]$reg.output$yreg)$weights)
##
## Naive coefficient estimates:
## (Intercept) A M C1_error C2 A:M
## -2.5688363 -0.8881518 -1.8387439 0.4856104 -0.6142071 1.1063464
##
## Naive var-cov estimates:
## (Intercept) A M C1_error C2
## (Intercept) 0.067990581 -0.0545603114 -0.048204212 -0.0154891419 -0.0071194416
## A -0.054560311 0.5747507824 0.054868704 -0.0004875592 0.0004879114
## M -0.048204212 0.0548687045 0.081668670 -0.0080216586 -0.0027107101
## C1_error -0.015489142 -0.0004875592 -0.008021659 0.0219233192 -0.0001013951
## C2 -0.007119442 0.0004879114 -0.002710710 -0.0001013951 0.0225065272
## A:M 0.055848645 -0.5747208888 -0.077864595 -0.0011835634 -0.0008171310
## A:M
## (Intercept) 0.055848645
## A -0.574720889
## M -0.077864595
## C1_error -0.001183563
## C2 -0.000817131
## A:M 0.606845225
##
## Variable measured with error:
## C1_error
## Measurement error:
## [1] 0.05
##
## Error-corrected results:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -2.5697 0.2612 -9.839 < 2e-16 ***
## A -0.8885 0.7581 -1.172 0.24125
## M -1.8396 0.2859 -6.435 1.30e-10 ***
## C1_error 0.4875 0.1497 3.255 0.00114 **
## C2 -0.6141 0.1500 -4.093 4.29e-05 ***
## A:M 1.1065 0.7790 1.420 0.15554
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## ### Mediator regressions:
## Call:
## simexreg(reg = getCall(x$sens[[1L]]$reg.output$mreg[[1L]])$reg,
## formula = M ~ A + C1_error + C2, data = getCall(x$sens[[1L]]$reg.output$mreg[[1L]])$data,
## MEvariable = "C1_error", MEvartype = "continuous", MEerror = 0.05,
## variance = TRUE, lambda = c(0.5, 1, 1.5, 2), B = 200, weights = getCall(x$sens[[1L]]$reg.output$mreg[[1L]])$weights)
##
## Naive coefficient estimates:
## (Intercept) A C1_error C2
## 0.9915629 2.1481051 1.4448263 0.6769050
##
## Naive var-cov estimates:
## (Intercept) A C1_error C2
## (Intercept) 0.013444822 -0.0040002083 -0.0092978270 -0.0070380421
## A -0.004000208 0.0216896845 -0.0008364503 0.0002395075
## C1_error -0.009297827 -0.0008364503 0.0142375505 0.0008566472
## C2 -0.007038042 0.0002395075 0.0008566472 0.0142371683
##
## Variable measured with error:
## C1_error
## Measurement error:
## [1] 0.05
##
## Error-corrected results:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.9816 0.1165 8.426 < 2e-16 ***
## A 2.1466 0.1473 14.572 < 2e-16 ***
## C1_error 1.4580 0.1205 12.102 < 2e-16 ***
## C2 0.6768 0.1194 5.670 1.47e-08 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
##
## ## Error-corrected causal effects on the risk ratio scale for measurement error 1:
## Estimate Std.error 95% CIL 95% CIU P.val
## Rcde 1.24357 0.22301 0.87503 1.767 0.224154
## Rpnde 1.01920 0.17645 0.72592 1.431 0.912545
## Rtnde 1.20915 0.21032 0.85985 1.700 0.274902
## Rpnie 0.80111 0.05410 0.70180 0.914 0.001024 **
## Rtnie 0.95042 0.06585 0.82975 1.089 0.462970
## Rte 0.96867 0.15569 0.70691 1.327 0.842992
## ERcde 0.18834 0.16647 -0.13794 0.515 0.257913
## ERintref -0.16914 0.09979 -0.36473 0.026 0.090093 .
## ERintmed 0.14836 0.08769 -0.02352 0.320 0.090694 .
## ERpnie -0.19889 0.05410 -0.30492 -0.093 0.000237 ***
## ERcde(prop) -6.01072 34.85624 -74.32769 62.306 0.863089
## ERintref(prop) 5.39808 26.78430 -47.09817 57.894 0.840277
## ERintmed(prop) -4.73472 23.48910 -50.77251 41.303 0.840252
## ERpnie(prop) 6.34736 31.57520 -55.53889 68.234 0.840680
## pm 1.61264 8.50887 -15.06443 18.290 0.849682
## int 0.66336 3.29679 -5.79824 7.125 0.840532
## pe 7.01072 34.85624 -61.30625 75.328 0.840595
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## ----------------------------------------------------------------
##
## (Rcde: controlled direct effect risk ratio; Rpnde: pure natural direct effect risk ratio; Rtnde: total natural direct effect risk ratio; Rpnie: pure natural indirect effect risk ratio; Rtnie: total natural indirect effect risk ratio; Rte: total effect risk ratio; ERcde: excess relative risk due to controlled direct effect; ERintref: excess relative risk due to reference interaction; ERintmed: excess relative risk due to mediated interaction; ERpnie: excess relative risk due to pure natural indirect effect; ERcde(prop): proportion ERcde; ERintref(prop): proportion ERintref; ERintmed(prop): proportion ERintmed; ERpnie(prop): proportion ERpnie; pm: overall proportion mediated; int: overall proportion attributable to interaction; pe: overall proportion eliminated)
##
## Relevant variable values:
## $a
## [1] 1
##
## $astar
## [1] 0
##
## $yval
## [1] "1"
##
## $mval
## $mval[[1]]
## [1] 1
##
##
## $basecval
## $basecval[[1]]
## [1] 0.996522
##
## $basecval[[2]]
## [1] 0.5936A Categorical Variable Measured with Error
Then, we assume \(A\) was measured with error. \(A\) is categorical, so only SIMEX can be used. The naive results obtained by fitting data with measurement error:
res_naive_cat <- cmest(data = data, model = "rb", outcome = "Y", exposure = "A_error",
mediator = "M", basec = c("C1", "C2"), EMint = TRUE,
mreg = list("logistic"), yreg = "logistic",
astar = 0, a = 1, mval = list(1),
estimation = "paramfunc", inference = "delta")
summary(res_naive_cat)## Causal Mediation Analysis
##
## # Outcome regression:
##
## Call:
## glm(formula = Y ~ A_error + M + A_error * M + C1 + C2, family = binomial(),
## data = getCall(x$reg.output$yreg)$data, weights = getCall(x$reg.output$yreg)$weights)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -0.5652 -0.2098 -0.1765 -0.1529 3.1640
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -2.5907 0.2727 -9.500 < 2e-16 ***
## A_error -0.5077 0.5703 -0.890 0.373318
## M -1.7375 0.2882 -6.029 1.65e-09 ***
## C1 0.5037 0.1487 3.386 0.000709 ***
## C2 -0.6133 0.1500 -4.089 4.34e-05 ***
## A_error:M 0.5931 0.5945 0.998 0.318446
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 1858.7 on 9999 degrees of freedom
## Residual deviance: 1801.3 on 9994 degrees of freedom
## AIC: 1813.3
##
## Number of Fisher Scoring iterations: 7
##
##
## # Mediator regressions:
##
## Call:
## glm(formula = M ~ A_error + C1 + C2, family = binomial(), data = getCall(x$reg.output$mreg[[1L]])$data,
## weights = getCall(x$reg.output$mreg[[1L]])$weights)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -3.3245 0.1157 0.1712 0.2666 1.0700
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 1.1369 0.1164 9.769 < 2e-16 ***
## A_error 1.5434 0.1310 11.783 < 2e-16 ***
## C1 1.5322 0.1190 12.879 < 2e-16 ***
## C2 0.6753 0.1181 5.718 1.08e-08 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 2825.7 on 9999 degrees of freedom
## Residual deviance: 2428.4 on 9996 degrees of freedom
## AIC: 2436.4
##
## Number of Fisher Scoring iterations: 7
##
##
## # Effect decomposition on the odds ratio scale via the regression-based approach
##
## Closed-form parameter function estimation with
## delta method standard errors, confidence intervals and p-values
##
## Estimate Std.error 95% CIL 95% CIU P.val
## Rcde 1.08912 0.18279 0.78381 1.513 0.611003
## Rpnde 0.98700 0.15917 0.71952 1.354 0.935309
## Rtnde 1.06299 0.17173 0.77449 1.459 0.705322
## Rpnie 0.86553 0.04070 0.78932 0.949 0.002133 **
## Rtnie 0.93218 0.04836 0.84205 1.032 0.175816
## Rte 0.92005 0.14238 0.67934 1.246 0.590294
## ERcde 0.07373 0.14960 -0.21949 0.367 0.622139
## ERintref -0.08673 0.08188 -0.24722 0.074 0.289503
## ERintmed 0.06753 0.06388 -0.05767 0.193 0.290433
## ERpnie -0.13447 0.04070 -0.21424 -0.055 0.000954 ***
## ERcde(prop) -0.92222 3.45691 -7.69764 5.853 0.789642
## ERintref(prop) 1.08489 2.06437 -2.96120 5.131 0.599215
## ERintmed(prop) -0.84468 1.60658 -3.99352 2.304 0.599050
## ERpnie(prop) 1.68201 3.03652 -4.26946 7.633 0.579628
## pm 0.83733 1.71924 -2.53231 4.207 0.626232
## int 0.24020 0.45912 -0.65966 1.140 0.600849
## pe 1.92222 3.45691 -4.85321 8.698 0.578176
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Rcde: controlled direct effect odds ratio; Rpnde: pure natural direct effect odds ratio; Rtnde: total natural direct effect odds ratio; Rpnie: pure natural indirect effect odds ratio; Rtnie: total natural indirect effect odds ratio; Rte: total effect odds ratio; ERcde: excess relative risk due to controlled direct effect; ERintref: excess relative risk due to reference interaction; ERintmed: excess relative risk due to mediated interaction; ERpnie: excess relative risk due to pure natural indirect effect; ERcde(prop): proportion ERcde; ERintref(prop): proportion ERintref; ERintmed(prop): proportion ERintmed; ERpnie(prop): proportion ERpnie; pm: overall proportion mediated; int: overall proportion attributable to interaction; pe: overall proportion eliminated)
##
## Relevant variable values:
## $a
## [1] 1
##
## $astar
## [1] 0
##
## $yval
## [1] "1"
##
## $mval
## $mval[[1]]
## [1] 1
##
##
## $basecval
## $basecval[[1]]
## [1] 0.9967315
##
## $basecval[[2]]
## [1] 0.5936The results corrected by SIMEX:
res_simex_cat <- cmsens(object = res_naive_cat, sens = "me", MEmethod = "simex",
MEvariable = "A_error", MEvartype = "cat", MEerror = list(mc))
summary(res_simex_cat)## Sensitivity Analysis For Measurement Error
##
## The variable measured with error: A_error
## Type of the variable measured with error: categorical
##
## # Measurement error 1:
## [,1] [,2]
## [1,] 0.9 0.1
## [2,] 0.1 0.9
##
## ## Error-corrected regressions for measurement error 1:
##
## ### Outcome regression:
## Call:
## simexreg(reg = getCall(x$sens[[1L]]$reg.output$yreg)$reg, formula = Y ~
## A_error + M + A_error * M + C1 + C2, data = getCall(x$sens[[1L]]$reg.output$yreg)$data,
## MEvariable = "A_error", MEvartype = "categorical", MEerror = c(0.9,
## 0.1, 0.1, 0.9), variance = TRUE, lambda = c(0.5, 1, 1.5,
## 2), B = 200, weights = getCall(x$sens[[1L]]$reg.output$yreg)$weights)
##
## Naive coefficient estimates:
## (Intercept) A_error M C1 C2 A_error:M
## -2.5906738 -0.5077290 -1.7374904 0.5036807 -0.6133152 0.5930976
##
## Naive var-cov estimates:
## (Intercept) A_error M C1 C2
## (Intercept) 0.074373396 -0.0608915941 -0.054071017 -1.576936e-02 -7.164751e-03
## A_error -0.060891594 0.3252487905 0.060788376 -1.130950e-04 5.120972e-04
## M -0.054071017 0.0607883762 0.083061875 -8.233291e-03 -2.664520e-03
## C1 -0.015769362 -0.0001130950 -0.008233291 2.212526e-02 -9.887795e-05
## C2 -0.007164751 0.0005120972 -0.002664520 -9.887795e-05 2.250262e-02
## A_error:M 0.062066763 -0.3252496367 -0.079058624 -1.368648e-03 -8.758372e-04
## A_error:M
## (Intercept) 0.0620667626
## A_error -0.3252496367
## M -0.0790586237
## C1 -0.0013686479
## C2 -0.0008758372
## A_error:M 0.3534194114
##
## Variable measured with error:
## A_error
## Measurement error:
## 0 1
## 0 0.9 0.1
## 1 0.1 0.9
##
## Error-corrected results:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -2.5706 0.2726 -9.429 < 2e-16 ***
## A_error1 -0.6697 0.7186 -0.932 0.351439
## M -1.7728 0.2976 -5.957 2.66e-09 ***
## C1 0.5014 0.1490 3.366 0.000764 ***
## C2 -0.6139 0.1500 -4.092 4.31e-05 ***
## A_error1:M 0.7803 0.7398 1.055 0.291608
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## ### Mediator regressions:
## Call:
## simexreg(reg = getCall(x$sens[[1L]]$reg.output$mreg[[1L]])$reg,
## formula = M ~ A_error + C1 + C2, data = getCall(x$sens[[1L]]$reg.output$mreg[[1L]])$data,
## MEvariable = "A_error", MEvartype = "categorical", MEerror = c(0.9,
## 0.1, 0.1, 0.9), variance = TRUE, lambda = c(0.5, 1, 1.5,
## 2), B = 200, weights = getCall(x$sens[[1L]]$reg.output$mreg[[1L]])$weights)
##
## Naive coefficient estimates:
## (Intercept) A_error C1 C2
## 1.1368565 1.5434255 1.5321997 0.6752983
##
## Naive var-cov estimates:
## (Intercept) A_error C1 C2
## (Intercept) 0.013541986 -0.0044464941 -0.0091359634 -0.0068278415
## A_error -0.004446494 0.0171570798 -0.0006625250 0.0001919242
## C1 -0.009135963 -0.0006625250 0.0141546164 0.0007819793
## C2 -0.006827841 0.0001919242 0.0007819793 0.0139496499
##
## Variable measured with error:
## A_error
## Measurement error:
## 0 1
## 0 0.9 0.1
## 1 0.1 0.9
##
## Error-corrected results:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.0152 0.1193 8.512 < 2e-16 ***
## A_error1 1.9701 0.1676 11.757 < 2e-16 ***
## C1 1.4739 0.1203 12.255 < 2e-16 ***
## C2 0.6761 0.1197 5.649 1.66e-08 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
##
## ## Error-corrected causal effects on the risk ratio scale for measurement error 1:
## Estimate Std.error 95% CIL 95% CIU P.val
## Rcde 1.11696 0.23958 0.73360 1.701 0.606087
## Rpnde 0.96730 0.20112 0.64354 1.454 0.872960
## Rtnde 1.09045 0.22744 0.72455 1.641 0.678042
## Rpnie 0.82467 0.05202 0.72876 0.933 0.002245 **
## Rtnie 0.92966 0.06727 0.80674 1.071 0.313472
## Rte 0.89926 0.17449 0.61478 1.315 0.584228
## ERcde 0.09294 0.18747 -0.27449 0.460 0.620044
## ERintref -0.12564 0.10510 -0.33164 0.080 0.231910
## ERintmed 0.10729 0.08991 -0.06894 0.284 0.232769
## ERpnie -0.17533 0.05202 -0.27729 -0.073 0.000751 ***
## ERcde(prop) -0.92262 3.41032 -7.60672 5.761 0.786746
## ERintref(prop) 1.24722 2.29278 -3.24654 5.741 0.586456
## ERintmed(prop) -1.06501 1.95713 -4.90092 2.771 0.586326
## ERpnie(prop) 1.74041 3.09718 -4.32996 7.811 0.574162
## pm 0.67540 1.48233 -2.22991 3.581 0.648653
## int 0.18221 0.33721 -0.47870 0.843 0.588946
## pe 1.92262 3.41032 -4.76148 8.607 0.572913
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## ----------------------------------------------------------------
##
## (Rcde: controlled direct effect risk ratio; Rpnde: pure natural direct effect risk ratio; Rtnde: total natural direct effect risk ratio; Rpnie: pure natural indirect effect risk ratio; Rtnie: total natural indirect effect risk ratio; Rte: total effect risk ratio; ERcde: excess relative risk due to controlled direct effect; ERintref: excess relative risk due to reference interaction; ERintmed: excess relative risk due to mediated interaction; ERpnie: excess relative risk due to pure natural indirect effect; ERcde(prop): proportion ERcde; ERintref(prop): proportion ERintref; ERintmed(prop): proportion ERintmed; ERpnie(prop): proportion ERpnie; pm: overall proportion mediated; int: overall proportion attributable to interaction; pe: overall proportion eliminated)
##
## Relevant variable values:
## $a
## [1] 1
##
## $astar
## [1] 0
##
## $yval
## [1] "1"
##
## $mval
## $mval[[1]]
## [1] 1
##
##
## $basecval
## $basecval[[1]]
## [1] 0.9967315
##
## $basecval[[2]]
## [1] 0.5936Compare the error-corrected results with the true results:
res_true <- cmest(data = data, model = "rb", outcome = "Y", exposure = "A",
mediator = "M", basec = c("C1", "C2"), EMint = TRUE,
mreg = list("logistic"), yreg = "logistic",
astar = 0, a = 1, mval = list(1),
estimation = "paramfunc", inference = "delta")
summary(res_true)## Causal Mediation Analysis
##
## # Outcome regression:
##
## Call:
## glm(formula = Y ~ A + M + A * M + C1 + C2, family = binomial(),
## data = getCall(x$reg.output$yreg)$data, weights = getCall(x$reg.output$yreg)$weights)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -0.5659 -0.2096 -0.1762 -0.1524 3.1928
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -2.5768 0.2611 -9.869 < 2e-16 ***
## A -0.8888 0.7581 -1.172 0.241026
## M -1.8429 0.2859 -6.447 1.14e-10 ***
## C1 0.4970 0.1489 3.337 0.000845 ***
## C2 -0.6144 0.1500 -4.095 4.22e-05 ***
## A:M 1.1064 0.7790 1.420 0.155512
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 1858.7 on 9999 degrees of freedom
## Residual deviance: 1799.2 on 9994 degrees of freedom
## AIC: 1811.2
##
## Number of Fisher Scoring iterations: 7
##
##
## # Mediator regressions:
##
## Call:
## glm(formula = M ~ A + C1 + C2, family = binomial(), data = getCall(x$reg.output$mreg[[1L]])$data,
## weights = getCall(x$reg.output$mreg[[1L]])$weights)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -3.4257 0.0947 0.1396 0.2558 1.0542
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 0.9817 0.1163 8.442 < 2e-16 ***
## A 2.1466 0.1473 14.574 < 2e-16 ***
## C1 1.4572 0.1200 12.145 < 2e-16 ***
## C2 0.6764 0.1193 5.668 1.44e-08 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 2825.7 on 9999 degrees of freedom
## Residual deviance: 2305.6 on 9996 degrees of freedom
## AIC: 2313.6
##
## Number of Fisher Scoring iterations: 7
##
##
## # Effect decomposition on the odds ratio scale via the regression-based approach
##
## Closed-form parameter function estimation with
## delta method standard errors, confidence intervals and p-values
##
## Estimate Std.error 95% CIL 95% CIU P.val
## Rcde 1.24309 0.22291 0.87472 1.767 0.224946
## Rpnde 1.01814 0.17635 0.72506 1.430 0.917328
## Rtnde 1.20855 0.21018 0.85947 1.699 0.276073
## Rpnie 0.80040 0.05419 0.70093 0.914 0.001009 **
## Rtnie 0.95009 0.06605 0.82906 1.089 0.461459
## Rte 0.96733 0.15543 0.70599 1.325 0.836210
## ERcde 0.18777 0.16621 -0.13800 0.514 0.258600
## ERintref -0.16963 0.10003 -0.36569 0.026 0.089947 .
## ERintmed 0.14878 0.08790 -0.02351 0.321 0.090547 .
## ERpnie -0.19960 0.05419 -0.30582 -0.093 0.000231 ***
## ERcde(prop) -5.74662 32.11420 -68.68930 57.196 0.857982
## ERintref(prop) 5.19141 24.66335 -43.14787 53.531 0.833285
## ERintmed(prop) -4.55337 21.62860 -46.94465 37.838 0.833257
## ERpnie(prop) 6.10859 29.09367 -50.91396 63.131 0.833697
## pm 1.55521 7.88784 -13.90468 17.015 0.843698
## int 0.63803 3.03635 -5.31311 6.589 0.833565
## pe 6.74662 32.11420 -56.19606 69.689 0.833604
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Rcde: controlled direct effect odds ratio; Rpnde: pure natural direct effect odds ratio; Rtnde: total natural direct effect odds ratio; Rpnie: pure natural indirect effect odds ratio; Rtnie: total natural indirect effect odds ratio; Rte: total effect odds ratio; ERcde: excess relative risk due to controlled direct effect; ERintref: excess relative risk due to reference interaction; ERintmed: excess relative risk due to mediated interaction; ERpnie: excess relative risk due to pure natural indirect effect; ERcde(prop): proportion ERcde; ERintref(prop): proportion ERintref; ERintmed(prop): proportion ERintmed; ERpnie(prop): proportion ERpnie; pm: overall proportion mediated; int: overall proportion attributable to interaction; pe: overall proportion eliminated)
##
## Relevant variable values:
## $a
## [1] 1
##
## $astar
## [1] 0
##
## $yval
## [1] "1"
##
## $mval
## $mval[[1]]
## [1] 1
##
##
## $basecval
## $basecval[[1]]
## [1] 0.9967315
##
## $basecval[[2]]
## [1] 0.5936